About Our Technology
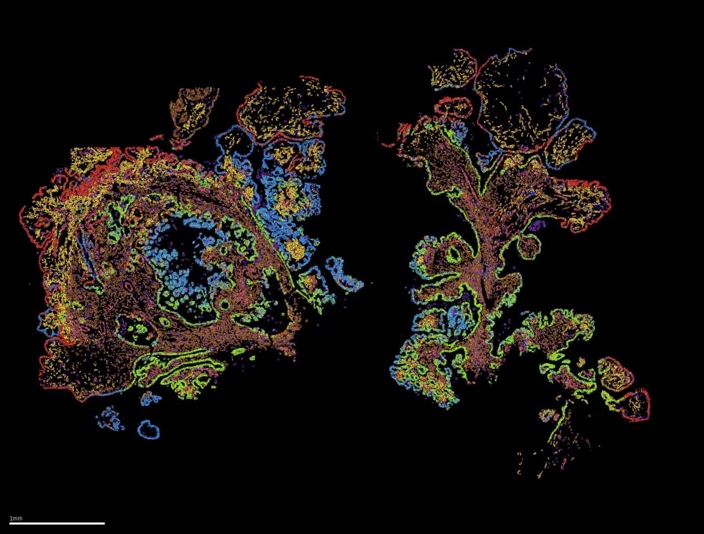
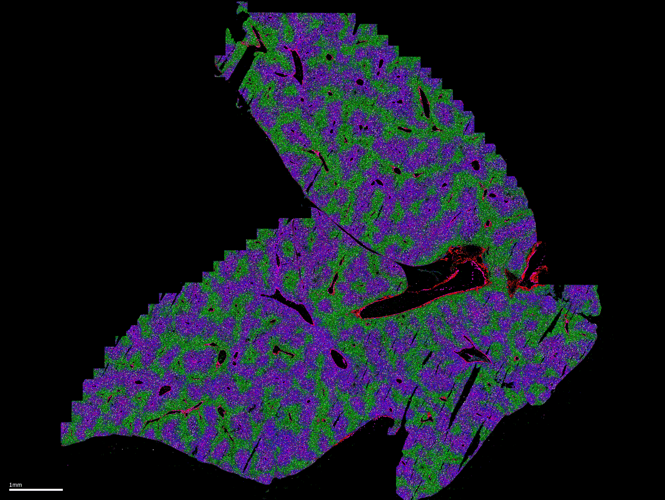
MERSCOPE Ultra™ from Vizgen is a next-generation spatial genomics platform designed for high-throughput, high-resolution RNA mapping across whole tissue sections. It delivers single-cell and subcellular precision, enabling researchers to visualize and quantify gene expression in situ. The system supports large-scale spatial transcriptomic studies with expanded imaging capacity and operational flexibility, thanks to two flow cell sizes – standard and large – allowing imaging of up to 9 cm² per week. This flexibility accommodates a wide range of experimental designs, from large tissue sections to multiple smaller samples, optimizing both efficiency and cost-effectiveness.
How Does MERSCOPE Ultra™ Work

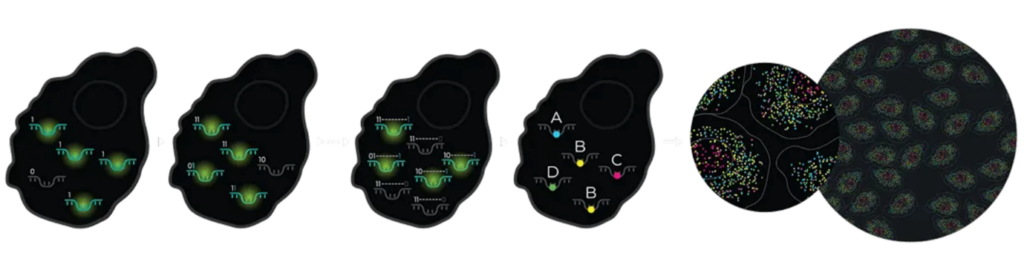
At the core of MERSCOPE Ultra™ is MERFISH (Multiplexed Error-Robust Fluorescence In Situ Hybridization), a spatial transcriptomics technique that enables direct detection and quantification of hundreds to thousands of RNA species within intact tissues. The process involves:
- Hybridizing barcoded oligonucleotide probe libraries directly to RNA molecules in situ.
- Sequential rounds of high-resolution imaging to decode transcript identity and spatial location.
- An error-robust barcoding strategy that ensures high detection accuracy, reproducibility, and scalability for single-cell spatial transcriptomic analysis.
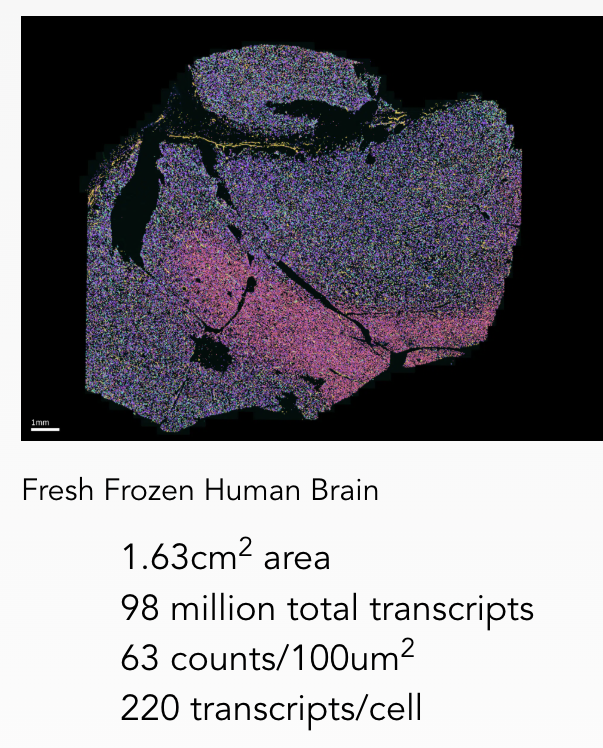
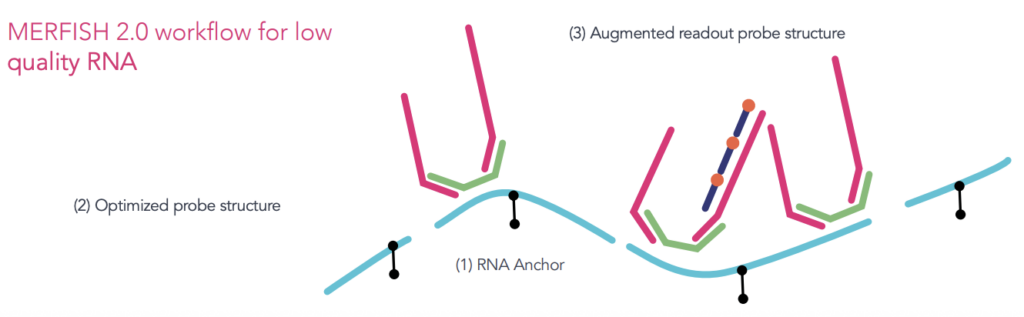
The latest MERFISH 2.0 chemistry introduces advanced RNA anchoring and improved signal amplification, increasing sensitivity and robustness, especially for degraded RNA samples such as those from formalin-fixed paraffin-embedded (FFPE) tissues. These improvements make it possible to generate high-quality spatial data from challenging specimen types such as formalin-fixed paraffin-embedded (FFPE) tissues, which are widely used in clinical and archival research but historically difficult to analyze due to RNA degradation. By combining the scale and flexibility of MERSCOPE Ultra™ with the heightened performance of MERFISH 2.0, researchers can now spatially profile gene expression across an even broader array of biological and pathological samples. This integrated platform is ideal for a deeper understanding of spatial gene regulation, cell-cell interactions, and tissue heterogeneity.
Why Choose MERSCOPE Ultra™
- High-Resolution, High-Throughput Spatial Transcriptomics
Capture spatial gene expression at single-cell and subcellular resolution with exceptional sensitivity. MERSCOPE Ultra™ enables comprehensive profiling of tissue architecture and cellular states, uncovering fine-scale heterogeneity and molecular interactions. - Unmatched Flexibility for Experimental Design
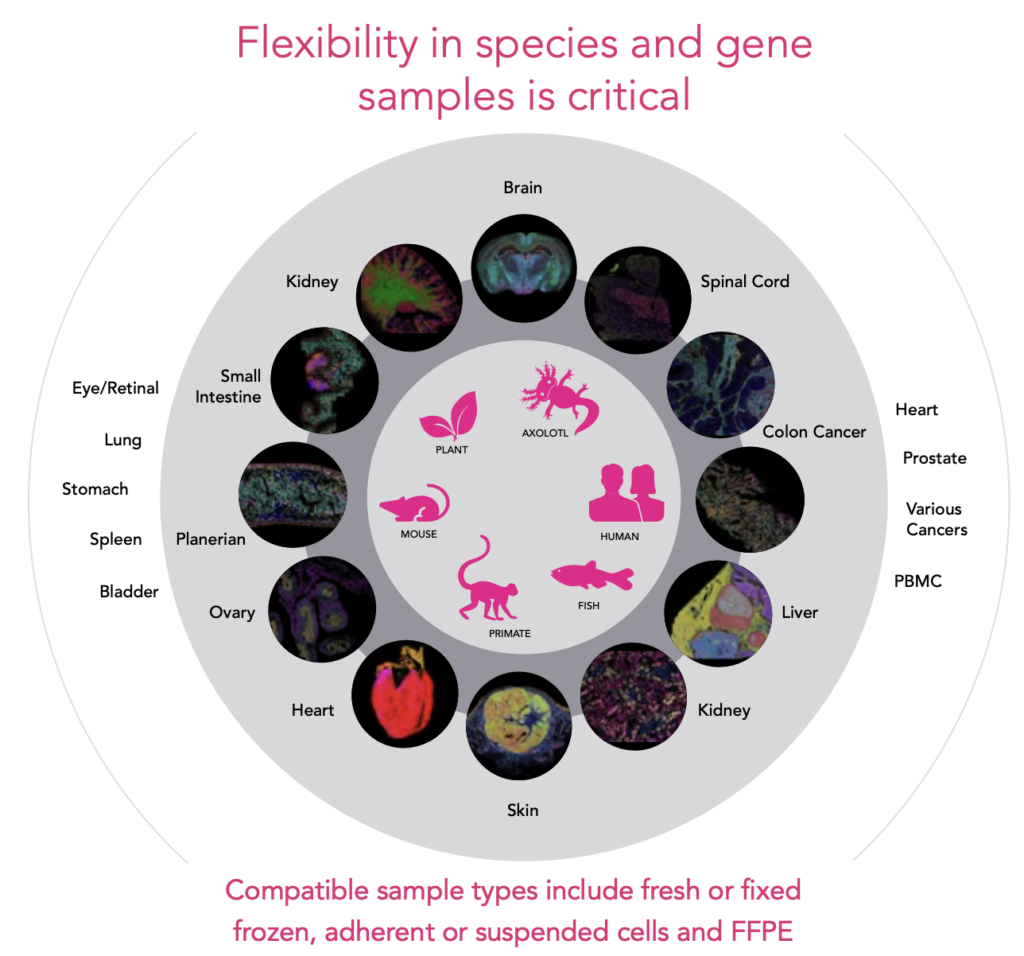
Accommodate large tissue sections or multiple smaller samples within a single run, thanks to a scalable imaging area of up to 3.0 cm². Customize panels targeting up to 1000 genes across diverse species and sample types – including FFPE, frozen, and adherent or suspended cells.
- Reliable Performance with MERFISH 2.0 Chemistry
Powered by advanced MERFISH 2.0 technology, MERSCOPE Ultra delivers robust results even from degraded or archival samples. Enhanced probe chemistry and transcript anchoring ensure high sensitivity, specificity, and accurate localization—ideal for challenging samples and high-plex applications.
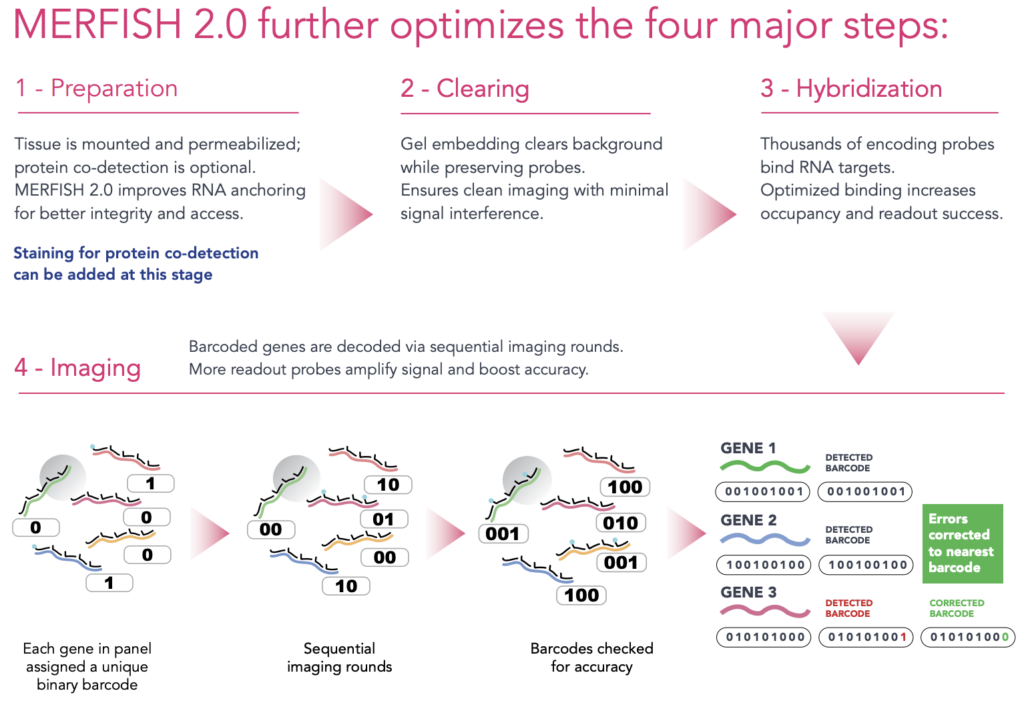
Workflow: From Sample to Spatial Insight
Gene Panel Options
MERSCOPE Ultra™ supports a range of high-plex gene panels designed for diverse research needs:
| Panel Type | Description |
|---|---|
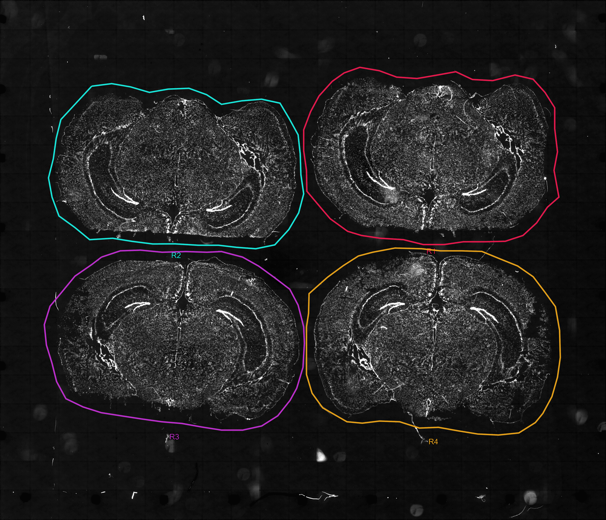
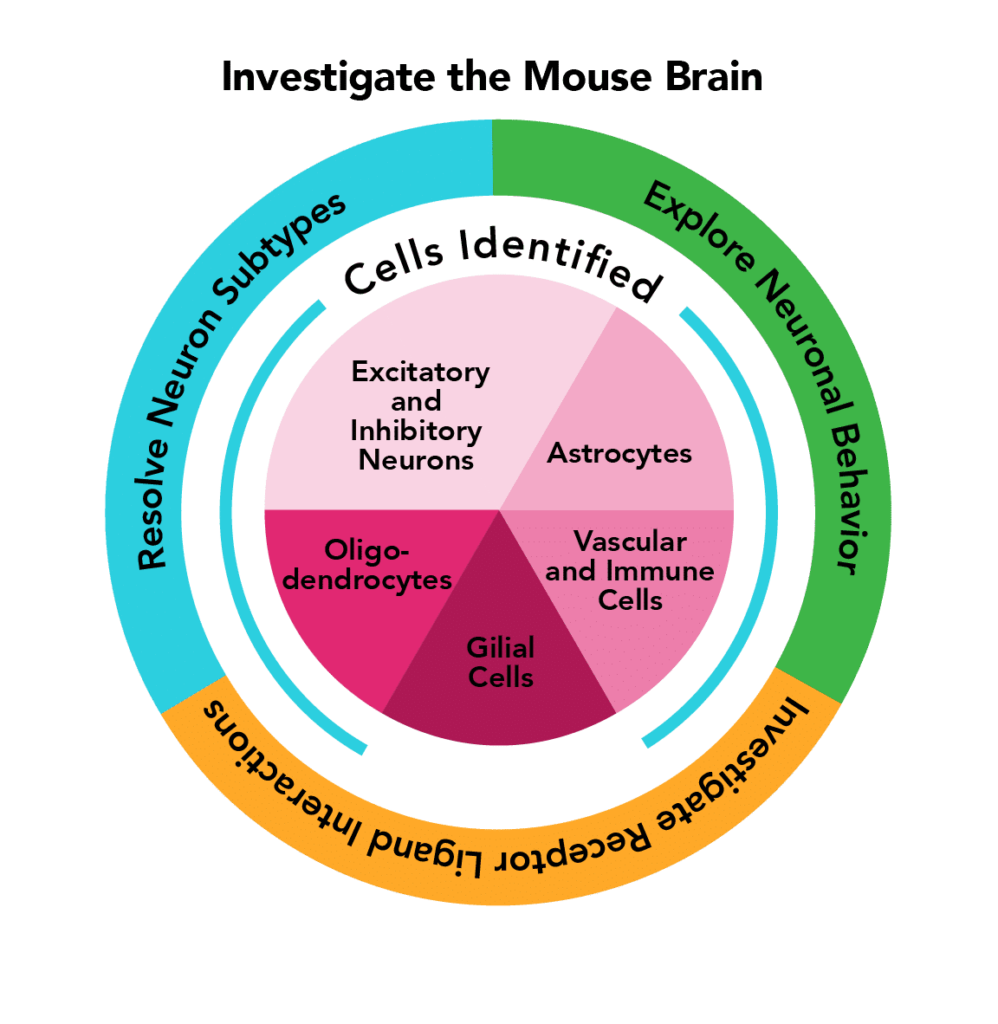
| PanNeuro Cell Type | MERSCOPE 500 gene predesigned panel providing the ability to cell type major cells (e.g. excitatory and inhibitory neurons, glial cells, astrocytes, oligodendrocytes and vascular cells) in mouse brain tissue samples. |
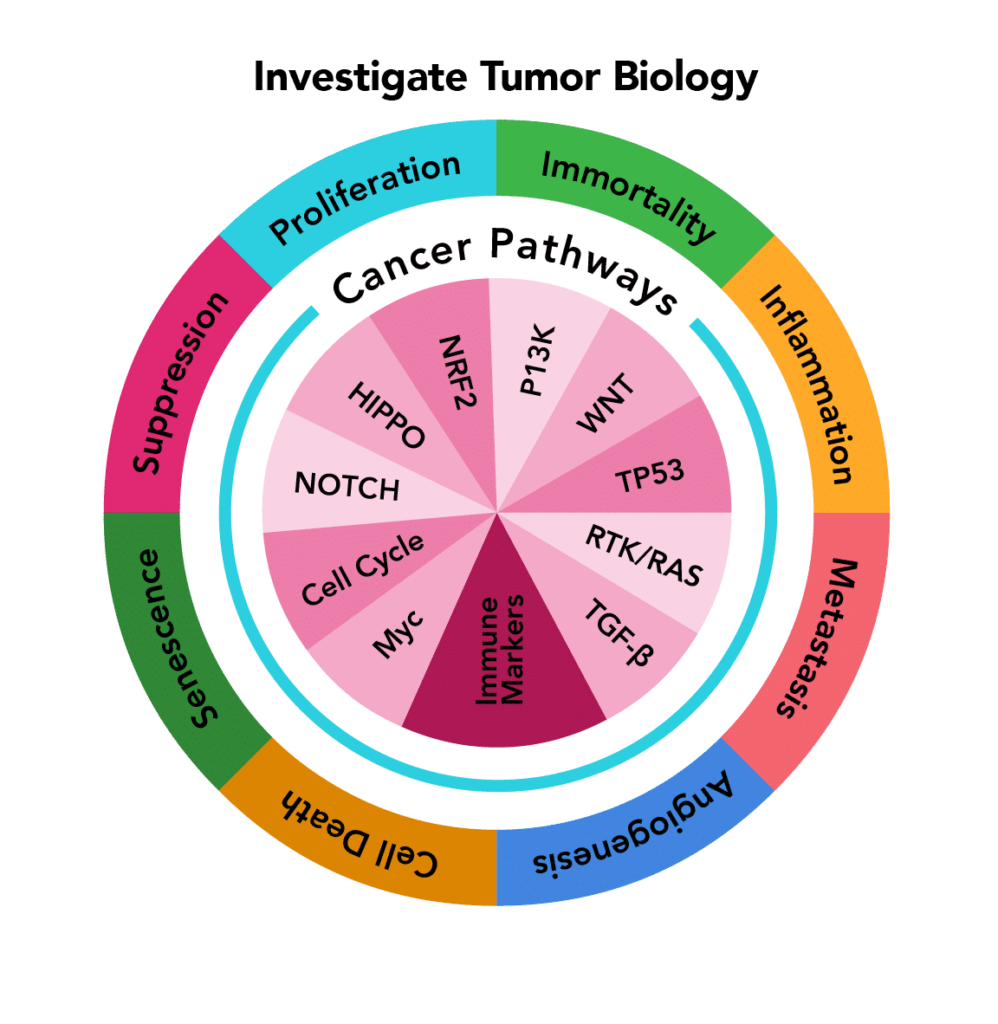
| PanCancer Pathways | MERSCOPE 500 gene predesigned panel targeting canonical signalling pathways in human cancer tissue samples. Illuminate the tumour microenvironment and understand oncological heterogeneity. |
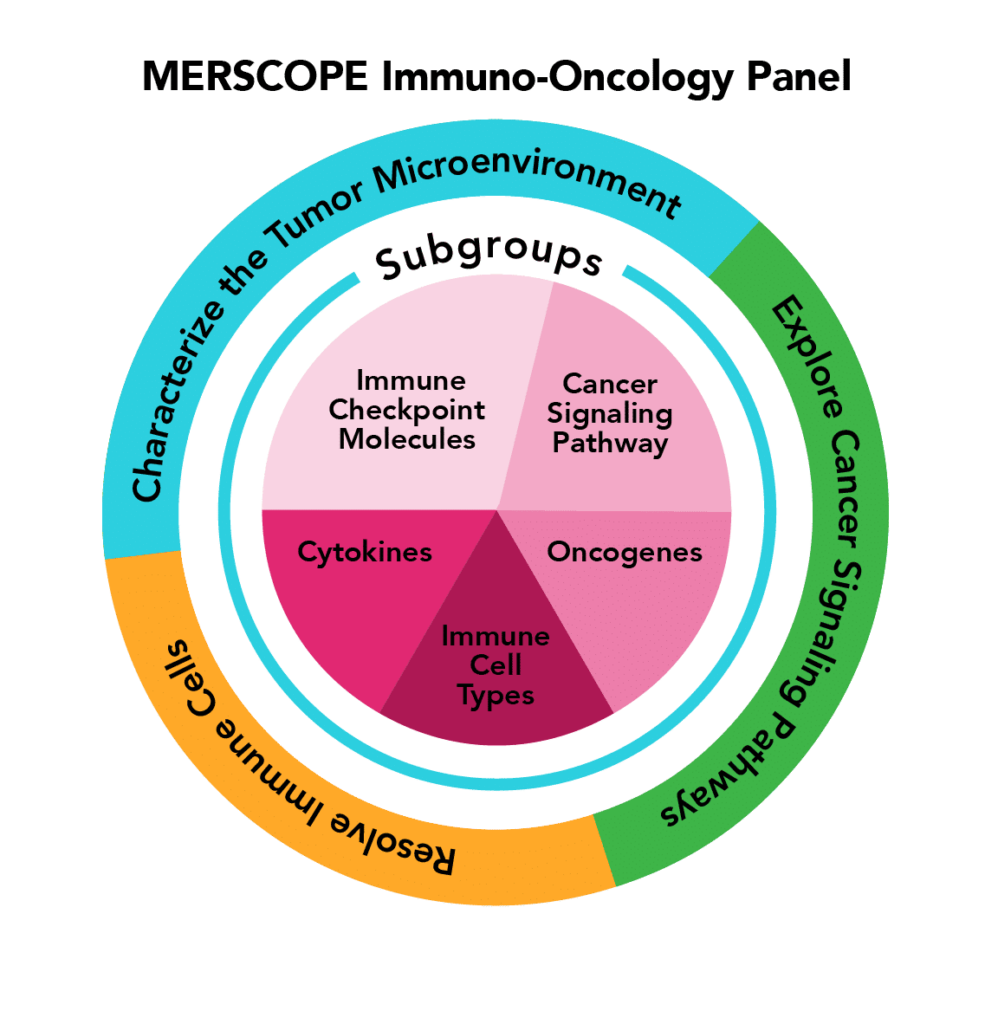
| Immuno-Oncology | MERSCOPE 500 gene predesigned panel allows for the characterization of tumour and immune behaviour at cellular and subcellular levels across multiple types of human cancers (Breast Cancer, Lung Cancer, Liver Cancer, Prostate Cancer, Colon Cancer, Melanoma, Ovarian Cancer, Brain Cancer, Uterine Cancer) |
| Custom Panel | MERSCOPE Custom Panels offer you the ability to build a panel with up to 1000 genes specifically chosen for your study. |
Applications
- Cell Atlas & Characterization: Define cell types, states, and spatial gene expression patterns across tissues.
- Tissue Microenvironments: Investigate spatial cellular neighborhoods, cell–cell interactions, and heterogeneity.
- Disease Pathology: Visualize molecular and cellular changes within preserved tissue architecture.
- Biomarker Discovery: Identify disease-related biomarkers with spatial resolution.
- Ligand–Receptor Interactions: Map spatial expression of signalling pathways.
Key Technical Specifications
| Feature | Specification |
|---|---|
| Throughput | Up to 9.0 cm² per week |
| Multiplex Capacity | Up to 1000 genes |
| Optical Resolution | ≤ 20 nm (oil immersion objective) |
| Lateral Resolution | 100nm pixel size |
| On-Instrument Storage | 58 TB |
| Automated Image Processing | Automated transcript decoding and cell segmentation |
| Sample Compatibility | Fresh/fixed frozen tissue, FFPE, adherent or suspended cells |
| Supported Species | Human, mouse, axolotl, zebrafish, primate, plant, and more |
Advanced Data Analysis & Integration
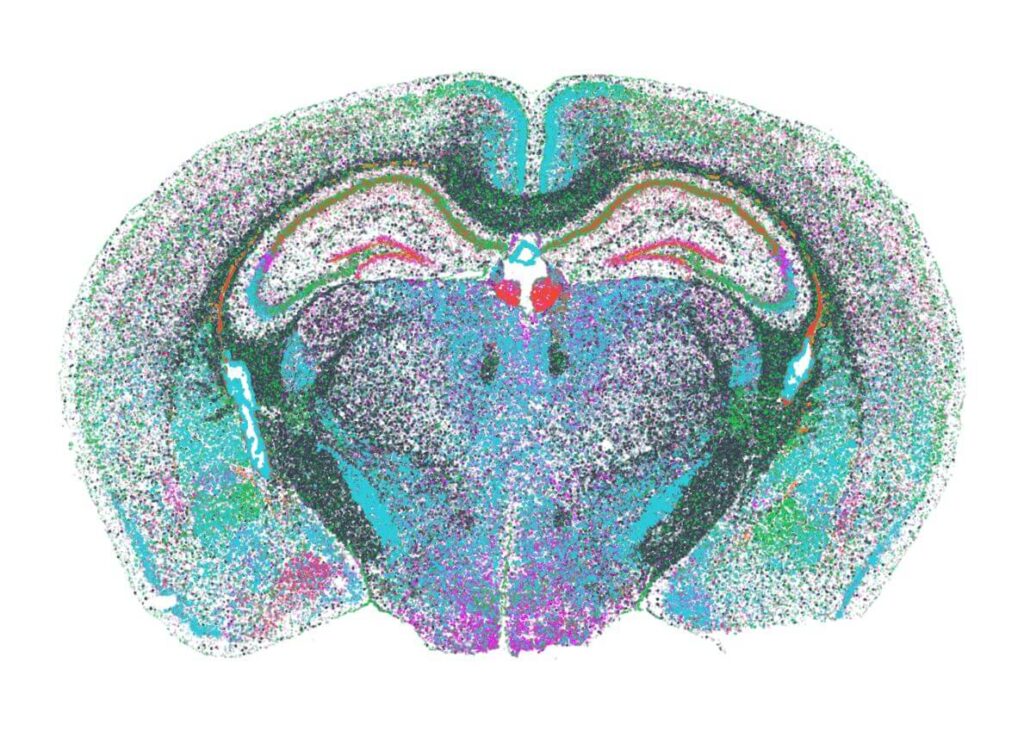
- High-Precision Segmentation: Integrated tools enable accurate cell segmentation, even in dense tissues, ensuring reliable transcript-to-cell assignment for downstream analysis.
- Third-Party Software Compatibility: Supports seamless data export to popular platforms like Seurat, Scanpy, BioTuring, and Heavy.AI for advanced single-cell and spatial transcriptomics analyses.
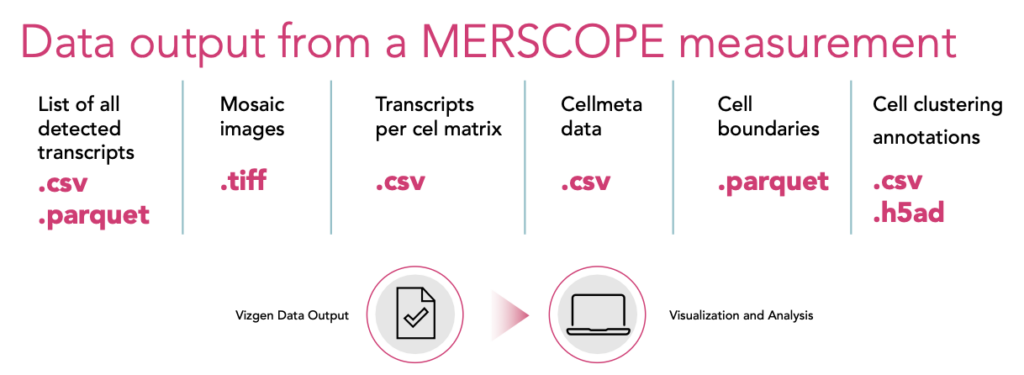
- Open-Source Post-Processing: The Vizgen Post Processing Tool (VPT) allows scalable, reproducible reprocessing of MERFISH data, supporting integration with other segmentation pipelines and deployment on workstations, clusters, or cloud environments.
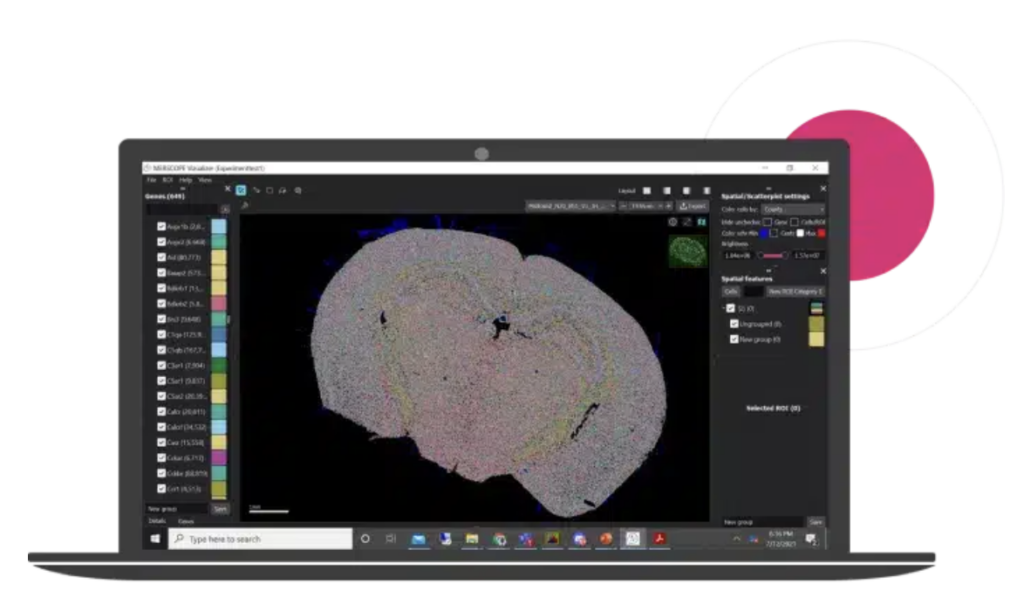
- MERSCOPE Visualizer: An interactive software for exploring spatial datasets, supporting .VZG, .CSV, .Parquet, and .HDF5 formats. Enables visualization of UMAP/t-SNE, Leiden clustering, region annotation, side-by-side dataset comparison, and high-resolution export.
Researchers can use these tools to interpret spatial gene expression, build cell atlases, and uncover tissue architecture with single-cell resolution.